
Computational Chemistry
The Department of Chemical Sciences hosts the Computational Chemistry Community at Padua / Comunità di Chimica Computazionale di Padova, or C3P.
In silico design or high-throughput computational approaches are actually familiar tasks in the scientific community activities. In addition to the theoretical method developers, chemists are by now computer science experts as they modify chemistry software to run on massively parallel machines with dozens, even hundreds, of processors, design general tools to make clusters and their ilk run better, highlight new research on ways to visualize data, design new hardware to speed up lengthy calculations, and test numerous parallel versions of computational chemistry software on new hardware architectures.
C3P aims to provide an efficient environment for chemists in Padova, utilizing complementary simulation and modelling tools, starting from static and dynamic ab initio quantum simulations (e.g. density functional theory), atomistic simulations based on detailed many-body inter-atomic potentials, steady-state and big-bang Monte Carlo methods, coarse-grained approaches based on molecular, brownian and dissipative particle dynamics and mesoscopic approaches. Besides C3P is a framework to develop general computational tools for nanoscience applications characterized by multiscale approaches and it is meant to be readily available for experimental research and for industrial applications of the specific computational tools developed in research academic environments.
The C3P main activities are devoted to computational and theoretical chemistry through a wide range of theoretical approaches and computational techniques, all focused on the interpretation, simulation, characterization and rationalization of complex molecular systems, functionalized materials at nanoscale, biomolecular processes.
C3P software repository
A repository of locally developed software is available for diverse chemical applications, under open source license.
In particular suites programs dedicated to magnetic spectroscopies are made available for the interpretation of relaxation properties from nuclear magnetic resonance (NMR) and simulation of electron spin resonance spectra (ESR). The link between theoretical predictive methodologies and MR spectroscopies dates back to several decades, and it is due to a happy coincidence between experimental needs and available interpretative tools. MR spectroscopies are intrinsically amenable to an advanced theoretical interpretation: the tools needed are based on quantum chemistry, as far as the parameters of the spin Hamiltonian are concerned, and on statistical thermodynamics, for the spectral lineshapes.
Presently, the main home-made available software packages are:
- E-SpiReS: an integrated approach to electron spin resonance;
- C++OPPS: a two-body stochastic approach to NMR relaxation in bio-molecules;
- DITE: evaluation of diffusion and friction tensors of flexible molecules;
- JEMDI: stochastic evaluation of multi-dimensional integrals;
- JEFREE: a C++ library for free-energy calculations within the framework of non-equilibrium transformations.
Commercial and open source software are also commonly running at C3P. Among others, the following tools are available: Adf, Amber, Comsol, Gaussian, Gromacs, Lammps, Mathematica, Materials Studio, Namd, Orca, Quantum espresso.
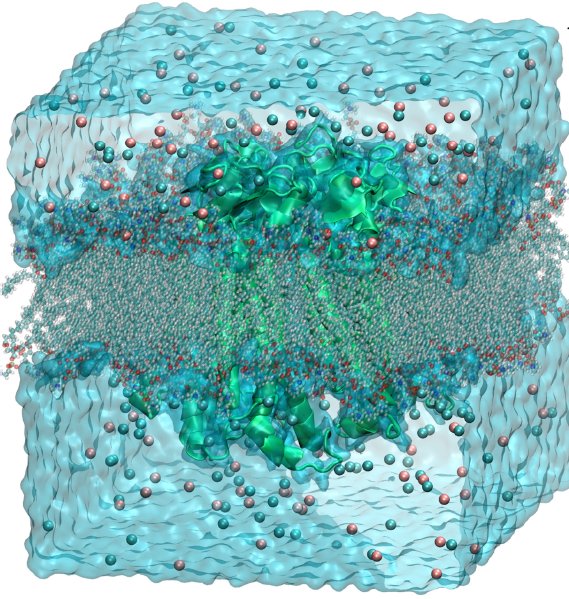
All-atom molecular dynamics simulation study of the mechanism, at atomic level, of the transport of Ca2+ ions through the channel of connexin. The latter is a trans-membrane protein consisting of about 10000 atoms. The simulations were carried out including a portion of phospholipidic bilayer, and explicit water molecules and calcium ions for a total of 100000 atoms.





